Handle Imbalanced Dataset
Handle Imbalanced Dataset
- (Along with Implementation in python!)

Let's take an example of Cancer Patient dataset where we are checking whether a person is having cancer or not based upon the input features.
Suppose in our dataset we have 1000 records and out of those 1000, 900 are the ones having cancer and rest 100 is non-cancer patient data.
So it is clearly an example of imbalance dataset as we have more number of rows with people having cancer than not having cancer. So if we train our data with this imbalance dataset and test it later with the new testing data, our model will be a lot partial towards the people having cancer as we have trained our model with the imbalanced dataset and thus our model accuracy with being very less in that case.
So How do we handle the Imbalance Dataset? Let look at some of the great techniques to avoid this kind of problem and train our model in a more precise way.
Now, we will use a couple of techniques to resolve this imbalanced dataset problem, lets look at those 2 techniques:
- Under Sampling
- Over Sampling
“Under Sampling”: One of the most common and simplest strategies to handle imbalanced data is to under-sample the majority class.
So, for this analysis, we will simply select n samples at random from the majority class, where n is the number of samples for the minority class, and use them during the training phase, after excluding the sample to use for validation. We will achieve this by using the NearMiss method of under-sampling.
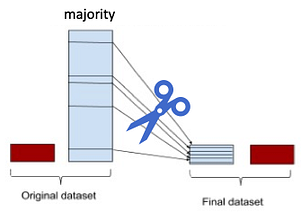
The one obvious disadvantage we have of using the under-sampling in that by under-sampling, we can solve the class imbalance issue but it will increase the sensitivity of our models. However, the results will be very poor. A reason could indeed be that we are training our classifiers using a few samples. In general, the more imbalanced the dataset the more samples will be discarded when under-sampling, therefore throwing away potentially useful information.
“Over Sampling”: The easiest way to oversample is to re-sample the minority class, i.e. to duplicate the entries or manufacture data which is exactly the same as what we have already.
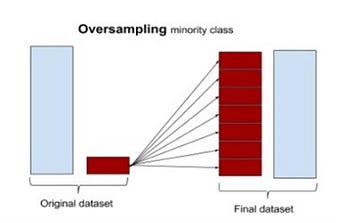
Oversampling the minority class can result in over-fitting problems if we over-sample before cross-validating. What is wrong with oversampling before cross-validating? Let’s consider the simplest oversampling method ever, as an example that clearly explains this point.
From left to right, we start with the original dataset where we have a minority class with two samples. We duplicate those samples, and then we do cross-validation. At this point there will be iterations, such as the one showed, where the training and validation set contains the same sample, resulting in over-fitting and misleading results.
Here is how this should be done:
First, we start cross-validating. This means that at each iteration we first exclude the sample to use as a validation set, and then over-sample the remaining of the minority class (in orange), this way we can avoid over-fitting our model.
This was a simple example, and better methods can be used to over-sample. One of the most common being the SMOTE technique, i.e. a method that instead of simply duplicating entries creates entries that are interpolations of the minority class, as well as under-samples the majority class.
We will be implementing all these “under-sampling” and “over-sampling” techniques below in python and will see the expected outcomes.
Let’s code now..!
Python Implementation:
Let’s take an example of Credit Card dataset where we have the output feature as “Fraud” or “not Fraud” for each and every transaction.
Step 1) Importing necessary libraries and the data:
import numpy as np
import pandas as pd
import sklearn
import scipy
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics import classification_report,accuracy_score
from sklearn.ensemble import IsolationForest
from sklearn.neighbors import LocalOutlierFactor
from sklearn.svm import OneClassSVM
from pylab import rcParams
rcParams[‘figure.figsize’] = 14, 8
RANDOM_SEED = 42
LABELS = [“Normal”, “Fraud”]#Importing the dataset
data = pd.read_csv(‘creditcard.csv’,sep=’,’)
Step 2) Create independent and Dependent Features:
columns = data.columns.tolist()
columns = [c for c in columns if c not in [“Class”]]
target = “Class”
state = np.random.RandomState(42)
X = data[columns]
Y = data[target]
Here in the dataset, we have around 284807 rows and 30 columns, you can check that with X.shape and Y.shape
Step 3) Exploratory Data Analysis:
count_classes = pd.value_counts(data[‘Class’], sort = True)
count_classes.plot(kind = ‘bar’, rot=0)
plt.title(“Transaction Class Distribution”)
plt.xticks(range(2), LABELS)
plt.xlabel(“Class”)
plt.ylabel(“Frequency”)
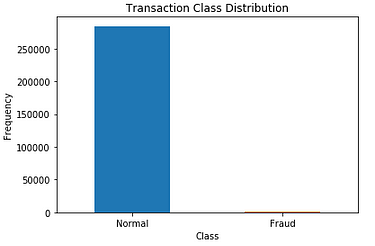
Output: You can see in the below graph that we have more than 2.5 lakh records having “Normal” transactions and very less number of records with “Fraud” transactions. So it is clearly a perfect example of the imbalanced dataset.
Step 4)Implementing Undersampling for Handling Imbalanced, Here we will be using “NearMiss” library of under_sampling:
from imblearn.under_sampling import NearMiss
nm = NearMiss(random_state=42)
X_res,y_res=nm.fit_sample(X,Y)
Step 5) Check the size of both the data now before and after applying the under-sampling method:
from collections import Counter
print(‘Original dataset shape {}’.format(Counter(Y)))
print(‘Resampled dataset shape {}’.format(Counter(y_res)))
Output:
Original dataset shape Counter({0: 284315, 1: 492})
Resampled dataset shape Counter({0: 492, 1: 492})
Just check above! we have under-sampled our data and now dataset is perfectly balanced for modelling.
Step 5)Implementing Oversampling for Handling Imbalanced, Here we are going to use “RandomOverSampler” and “SMOTETomek” library of Oversampling:
## RandomOverSampler to handle imbalanced data
from imblearn.over_sampling import RandomOverSampler
os = RandomOverSampler(ratio=0.5)
##Fitting the data now:
X_train_res, y_train_res = os.fit_sample(X, Y)
That’s it! now you can check the shape of both X and Y like below:
X_train_res.shape,y_train_res.shape
Now we will use the SMOTETomek which is a method of imblearn. SMOTETomek is a hybrid method which uses an under-sampling method (Tomek) in with an oversampling method (SMOTE). The best method to use and t to get more accurate results.
os_us = SMOTETomek(ratio=0.5)
X_train_res1, y_train_res1 = os_us.fit_sample(X, Y)
it’s done! now you can check the shape of the both X and Y train:
X_train_res.shape,y_train_res.shape
So, SMOTETomek performs better than the other over-sampling methods because it instead of simply duplicating entries creates entries that are interpolations of the minority class, as well as under-samples the majority class.
Conclusion:
In this story we have seen how can we handle the imbalanced dataset and, with the main goal of understanding how to properly cross-validate when oversampling is used. Hopefully, it is now clear how to make your data which is imbalanced to the one which is more balance using under-sampling and oversampling methods. I hope it will be clear now after this story.
Thanks...




Comments
Post a Comment